Copyright 2014 Sebastian Raschka
smilite
smilite is a Python 3 module to download and analyze SMILE strings (Simplified Molecular-Input Line-entry System) of chemical compounds from ZINC (a free database of commercially-available compounds for virtual screening, http://zinc.docking.org)
Sections
Installation
You can use the following command to install PyPrind:
pip install smilite
or
easy_install smilite
Alternatively, you download the package manually from the Python Package Index https://pypi.python.org/pypi/smilite, unzip it, navigate into the package, and use the command:
python3 setup.py install
Documentation
After you installed the smilite module, you can import it in Python via import smilite.
The current functions include:
def get_zinc_smile(zinc_id):
Gets the corresponding SMILE string for a ZINC ID query from
the ZINC online database. Requires an internet connection.
Keyword arguments:
zinc_id (str): A valid ZINC ID, e.g. 'ZINC00029323'
Returns the SMILE string for the corresponding ZINC ID.
E.g., 'COc1cccc(c1)NC(=O)c2cccnc2'
def generate_zincid_smile_csv(zincid_list, out_file):
Generates a CSV file of ZINC_ID,SMILE_string entries by querying the ZINC online
database.
Keyword arguments:
zincid_list (str): Path to a UTF-8 or ASCII formatted file
that contains 1 ZINC_ID per row. E.g.,
ZINC0000123456
ZINC0000234567
[...]
out_file (str): Path to a new output CSV file that will be written.
print_prgress_bar (bool): Prints a progress bar to the screen if True.
def check_duplicate_smiles(zincid_list, out_file, compare_simplified_smiles=False,
print_progress_bar=False):
Scans a ZINC_ID,SMILE_string CSV file for duplicate SMILE strings.
Keyword arguments:
zincid_list (str): Path to a UTF-8 or ASCII formatted file that
contains 1 ZINC_ID per row.
E.g.,
ZINC12345678,Cc1ccc(cc1C)OCCOc2c(cc(cc2I)/C=N/n3cnnc3)OC
ZINC01234567,C[C@H]1CCCC[NH+]1CC#CC(c2ccccc2)(c3ccccc3)O
[...]
out_file (str): Path to a new output CSV file that will be written.
compare_simplified_smiles (bool): If true, SMILE strings will be simplified
for the comparison.
def simplify_smile(smile_str):
Simplifies a SMILE string by removing hydrogen atoms (H),
chiral specifications ('@'), charges (+ / -), '#'-characters,
and square brackets ('[', ']').
Keyword Arguments:
smile_str (str): A smile string, e.g., CC@HNCCS(=O)(=O)[O-])
Returns a simplified SMILE string, e.g., CC(CCC(=O)NCCS(=O)(=O)O)
Command Line Scripts Examples
If you downloaded the smilite package from https://pypi.python.org/pypi/smilite or https://github.com/rasbt/smilite, you can use the command line scripts I provide in the scripts/ dir.
gen_zincid_smile_csv.py
Generates a ZINC_ID,SMILE_STR csv file from a input file of ZINC IDs. The input file should consist of 1 columns with 1 ZINC ID per row.
Usage:
[shell]>> python3 gen_zincid_smile_csv.py in.csv out.csv
Example:
[shell]>> python3 gen_zincid_smile_csv.py ../examples/zinc_ids.csv ../examples/zid_smiles.csv
Screen Output:
Downloading SMILES 0% 100% [########## ] | ETA[sec]: 106.525
Input example file format:
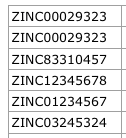
zinc_ids.csv
Output example file format:

zid_smiles.csv
comp_smile_strings.py
Compares SMILE strings in a 2 column CSV file (ZINC_ID,SMILE_string) to identify duplicates. Generates a new CSV file with ZINC IDs of identified duplicates listed in a 3rd-nth column(s).
Usage:
[shell]>> python3 comp_smile_strings.py in.csv out.csv [simplify]
Example 1:
[shell]>> python3 gen_zincid_smile_csv.py ../examples/zinc_ids.csv ../examples/zid_smiles.csv
Input example file format:

zid_smiles.csv
Output example file format 1:

comp_smiles.csv
Where
- 1st column: ZINC ID
- 2nd column: SMILE string
- 3rd column: number of duplicates
- 4th-nth column: ZINC IDs of duplicates
Example 2:
[shell]>> python3 comp_smile_strings.py ../examples/zid_smiles.csv ../examples/comp_simple_smiles.csv simplify
Output example file format 2:

comp_simple_smiles.csv
Contact
If you have any questions or comments about PyPrind, please feel free to contact me via
eMail: se.raschka@gmail.com
or Twitter: @rasbt
Changelog
VERSION 1.1.1
- PyPrind dependency fix
VERSION 1.1.0
- added a progress bar (pyprind) to
generate_zincid_smile_csv()function
smilite ======= smilite is a Python 3 module to download and analyze SMILE strings (Simplified Molecular-Input Line-entry System) of chemical compounds from ZINC (a free database of commercially-available compounds for virtual screening, [http://zinc.docking.org](http://zinc.docking.org)) ####Sections Installation ============= You can use the following command to install PyPrind: `pip install smilite` or `easy_install smilite` Alternatively, you download the package manually from the Python Package Index [https://pypi.python.org/pypi/smilite](https://pypi.python.org/pypi/smilite), unzip it, navigate into the package, and use the command: `python3 setup.py install`
Documentation ============= After you installed the smilite module, you can import it in Python via `import smilite`. The current functions include:
def get_zinc_smile(zinc_id):
Gets the corresponding SMILE string for a ZINC ID query from
the ZINC online database. Requires an internet connection.
Keyword arguments:
zinc_id (str): A valid ZINC ID, e.g. 'ZINC00029323'
Returns the SMILE string for the corresponding ZINC ID.
E.g., 'COc1cccc(c1)NC(=O)c2cccnc2'
def generate_zincid_smile_csv(zincid_list, out_file):
Generates a CSV file of ZINC_ID,SMILE_string entries by querying the ZINC online
database.
Keyword arguments:
zincid_list (str): Path to a UTF-8 or ASCII formatted file
that contains 1 ZINC_ID per row. E.g.,
ZINC0000123456
ZINC0000234567
[...]
out_file (str): Path to a new output CSV file that will be written.
print_prgress_bar (bool): Prints a progress bar to the screen if True.
def check_duplicate_smiles(zincid_list, out_file, compare_simplified_smiles=False,
print_progress_bar=False):
Scans a ZINC_ID,SMILE_string CSV file for duplicate SMILE strings.
Keyword arguments:
zincid_list (str): Path to a UTF-8 or ASCII formatted file that
contains 1 ZINC_ID per row.
E.g.,
ZINC12345678,Cc1ccc(cc1C)OCCOc2c(cc(cc2I)/C=N/n3cnnc3)OC
ZINC01234567,C[C@H]1CCCC[NH+]1CC#CC(c2ccccc2)(c3ccccc3)O
[...]
out_file (str): Path to a new output CSV file that will be written.
compare_simplified_smiles (bool): If true, SMILE strings will be simplified
for the comparison.
def simplify_smile(smile_str):
Simplifies a SMILE string by removing hydrogen atoms (H),
chiral specifications ('@'), charges (+ / -), '#'-characters,
and square brackets ('[', ']').
Keyword Arguments:
smile_str (str): A smile string, e.g., C[C@H](CCC(=O)NCCS(=O)(=O)[O-])
Returns a simplified SMILE string, e.g., CC(CCC(=O)NCCS(=O)(=O)O)
Command Line Scripts Examples ============= If you downloaded the smilite package from [https://pypi.python.org/pypi/smilite](https://pypi.python.org/pypi/smilite) or [https://github.com/rasbt/smilite](https://github.com/rasbt/smilite), you can use the command line scripts I provide in the `scripts/` dir.
###gen_zincid_smile_csv.py Generates a ZINC_ID,SMILE_STR csv file from a input file of ZINC IDs. The input file should consist of 1 columns with 1 ZINC ID per row. **Usage:** `[shell]>> python3 gen_zincid_smile_csv.py in.csv out.csv` **Example:** `[shell]>> python3 gen_zincid_smile_csv.py ../examples/zinc_ids.csv ../examples/zid_smiles.csv` **Screen Output:**
Downloading SMILES 0% 100% [########## ] | ETA[sec]: 106.525
**Input example file format:**  [zinc_ids.csv](https://raw.github.com/rasbt/smilite/master/examples/zinc_ids.csv)
**Output example file format:**  [zid_smiles.csv](https://raw.github.com/rasbt/smilite/master/examples/zid_smiles.csv)
###comp_smile_strings.py Compares SMILE strings in a 2 column CSV file (ZINC_ID,SMILE_string) to identify duplicates. Generates a new CSV file with ZINC IDs of identified duplicates listed in a 3rd-nth column(s). **Usage:** `[shell]>> python3 comp_smile_strings.py in.csv out.csv [simplify]` **Example 1:** `[shell]>> python3 gen_zincid_smile_csv.py ../examples/zinc_ids.csv ../examples/zid_smiles.csv`
**Input example file format:**  [zid_smiles.csv](https://raw.github.com/rasbt/smilite/master/examples/zid_smiles.csv)
**Output example file format 1:**  [comp_smiles.csv](https://raw.github.com/rasbt/smilite/master/examples/comp_smiles.csv)
Where - 1st column: ZINC ID - 2nd column: SMILE string - 3rd column: number of duplicates - 4th-nth column: ZINC IDs of duplicates
**Example 2:** `[shell]>> python3 comp_smile_strings.py ../examples/zid_smiles.csv ../examples/comp_simple_smiles.csv simplify`
**Output example file format 2:**  [comp_simple_smiles.csv](https://raw.github.com/rasbt/smilite/master/examples/comp_simple_smiles.csv)
Contact ============= If you have any questions or comments about PyPrind, please feel free to contact me via eMail: [se.raschka@gmail.com](mailto:se.raschka@gmail.com) or Twitter: [@rasbt](https://twitter.com/rasbt)
Changelog ========== **VERSION 1.1.1** - PyPrind dependency fix **VERSION 1.1.0** - added a progress bar (pyprind) to `generate_zincid_smile_csv()` function ��������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������smilite-1.1.1/scripts/������������������������������������������������������������������������������0000755�0000766�0000024�00000000000�12305163047�015745� 5����������������������������������������������������������������������������������������������������ustar �sebastian�����������������������staff���������������������������0000000�0000000������������������������������������������������������������������������������������������������������������������������������������������������������������������������smilite-1.1.1/scripts/comp_smile_strings.py���������������������������������������������������������0000644�0000766�0000024�00000003760�12305162131�022216� 0����������������������������������������������������������������������������������������������������ustar �sebastian�����������������������staff���������������������������0000000�0000000������������������������������������������������������������������������������������������������������������������������������������������������������������������������# Copyright 2014 Sebastian Raschka # # Compares SMILE strings in a 2 column CSV file (ZINC_ID,SMILE_string) to # identify duplicates. Generates a new CSV file with ZINC IDs of identified # duplicates listed in a 3rd-nth column(s). # # # Input example file format: # ZINC00029323,COc1cccc(c1)NC(=O)c2cccnc2 # ZINC00029323,COc1cccc(c1)NC(=O)c2cccnc2 # ZINC83310457,Cc1cccc(c1n2c(cc(c2C)/C=N\NC(=O)C(=O)Nc3ccc(cc3)[N+](=O)[O-])C)C # ZINC01234567,C[C@H]1CCCC[NH+]1CC#CC(c2ccccc2)(c3ccccc3)O # # Output example file format: # zinc_id,smile_str,duplicates # ZINC00029323,COc1cccc(c1)NC(=O)c2cccnc2,1,ZINC00029323, # ZINC83310457,Cc1cccc(c1n2c(cc(c2C)/C=N\NC(=O)C(=O)Nc3ccc(cc3)[N+](=O)[O-])C)C,0, # ZINC01234567,C[C@H]1CCCC[NH+]1CC#CC(c2ccccc2)(c3ccccc3)O,0, # # Where # 1st column: ZINC ID # 2nd column: SMILE string # 3rd column: number of duplicates # 4th-nth column: ZINC IDs of duplicates # # Usage: # [shell]>> python3 comp_smile_strings.py in.csv out.csv [simplify] # # Example1: # [shell]>> python3 gen_zincid_smile_csv.py ../examples/zinc_ids.csv ../examples/zid_smiles.csv # # Example2: # [shell]>> python3 comp_smile_strings.py ../examples/zid_smiles.csv ../examples/comp_simple_smiles.csv simplify import smilite import sys def print_usage(): print('\nUSAGE: python3 comp_smile_strings.py in.csv out.csv [simplify]') print('\nEXAMPLE1: python3 comp_smile_strings.py ../examples/zid_smiles.csv ../examples/comp_smiles.csv\n') print('\nEXAMPLE2: python3 comp_smile_strings.py ../examples/zid_smiles.csv ../examples/comp_simple_smiles.csv simplify\n') try: in_csv = sys.argv[1] out_csv = sys.argv[2] simplify = False if len(sys.argv) > 3: simplify = True smilite.check_duplicate_smiles(in_csv, out_csv, compare_simplified_smiles=simplify) except IOError as err: print('\n\nERROR: {}'.format(err)) print_usage() except IndexError: print('\n\nERROR: Invalid command line arguments.') print_usage() ����������������smilite-1.1.1/scripts/gen_zincid_smile_csv.py�������������������������������������������������������0000644�0000766�0000024�00000002211�12305162131�022461� 0����������������������������������������������������������������������������������������������������ustar �sebastian�����������������������staff���������������������������0000000�0000000������������������������������������������������������������������������������������������������������������������������������������������������������������������������# Copyright 2014 Sebastian Raschka # # Generates a ZINC_ID,SMILE_STR csv file from a input file of # ZINC IDs. The input file should consist of 1 columns with 1 ZINC ID per row. # # Input example file format: # ZINC0000123456 # ZINC0000234567 # ... # # Output example file format: # ZINC12345678,Cc1ccc(cc1C)OCCOc2c(cc(cc2I)/C=N/n3cnnc3)OC # ZINC01234567,C[C@H]1CCCC[NH+]1CC#CC(c2ccccc2)(c3ccccc3)O # ... # # Usage: # [shell]>> python3 gen_zincid_smile_csv.py in.csv out.csv # # Example: # [shell]>> python3 gen_zincid_smile_csv.py ../examples/zinc_ids.csv ../examples/zid_smiles.csv import smilite import sys def print_usage(): print('\nUSAGE: python3 gen_zincid_smile_csv.py in.csv out.csv') print('\nEXAMPLE: python3 gen_zincid_smile_csv.py ../examples/zinc_ids.csv ../examples/zid_smiles.csv\n') try: in_csv = sys.argv[1] out_csv = sys.argv[2] smilite.generate_zincid_smile_csv(in_csv, out_csv, print_progress_bar=True) except IOError as err: print('\n\nERROR: {}'.format(err)) print_usage() except IndexError: print('\n\nERROR: Invalid command line arguments.') print_usage() ���������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������smilite-1.1.1/setup.py������������������������������������������������������������������������������0000644�0000766�0000024�00000003553�12305162557�016003� 0����������������������������������������������������������������������������������������������������ustar �sebastian�����������������������staff���������������������������0000000�0000000������������������������������������������������������������������������������������������������������������������������������������������������������������������������from distutils.core import setup setup(name='smilite', version='1.1.1', description='smilite is a Python 3 module to download and analyze SMILE strings', author='Sebastian Raschka', author_email='se.raschka@gmail.com', url='https://github.com/rasbt/smilite', packages=['smilite'], data_files = [('', ['LICENSE.txt']), ('', ['README.html']), ('', ['README.md']), ('', ['CHANGELOG.txt']), ('test', ['test/test_get_zinc_smile.py']), ('test', ['test/test_simplify_smile.py']), ('examples', ['examples/comp_simple_smiles.csv']), ('examples', ['examples/comp_smiles.csv']), ('examples', ['examples/README.md']), ('examples', ['examples/zid_smiles.csv']), ('examples', ['examples/zinc_ids.csv']), ('scripts', ['scripts/comp_smile_strings.py']), ('scripts', ['scripts/gen_zincid_smile_csv.py']), ], install_requires=['PyPrind>=2.3.1'], license='GPLv3', platforms='any', classifiers=[ 'License :: OSI Approved :: GNU General Public License v3 (GPLv3)', 'Development Status :: 5 - Production/Stable', 'Programming Language :: Python :: 3', 'Environment :: Console', ], long_description=""" smilite is a Python 3 module to download and analyze SMILE strings (Simplified Molecular-Input Line-entry System) of chemical compounds from ZINC (a free database of commercially-available compounds for virtual screening, http://zinc.docking.org) Contact ============= If you have any questions or comments about PyPrind, please feel free to contact me via eMail: se.raschka@gmail.com or Twitter: https://twitter.com/rasbt """, ) �����������������������������������������������������������������������������������������������������������������������������������������������������smilite-1.1.1/smilite/������������������������������������������������������������������������������0000755�0000766�0000024�00000000000�12305163047�015724� 5����������������������������������������������������������������������������������������������������ustar �sebastian�����������������������staff���������������������������0000000�0000000������������������������������������������������������������������������������������������������������������������������������������������������������������������������smilite-1.1.1/smilite/__init__.py�������������������������������������������������������������������0000644�0000766�0000024�00000000571�12305163007�020034� 0����������������������������������������������������������������������������������������������������ustar �sebastian�����������������������staff���������������������������0000000�0000000������������������������������������������������������������������������������������������������������������������������������������������������������������������������# Copyright 2014 Sebastian Raschka # # A small module to retrieve SMILE strings # (Simplified molecular-input line-entry system) from the ZINC online # database (http://zinc.docking.org) from .smilite import get_zinc_smile from .smilite import generate_zincid_smile_csv from .smilite import simplify_smile from .smilite import check_duplicate_smiles __version__ = '1.1.1' ���������������������������������������������������������������������������������������������������������������������������������������smilite-1.1.1/smilite/smilite.py��������������������������������������������������������������������0000644�0000766�0000024�00000010576�12305162131�017746� 0����������������������������������������������������������������������������������������������������ustar �sebastian�����������������������staff���������������������������0000000�0000000������������������������������������������������������������������������������������������������������������������������������������������������������������������������# Copyright 2014 Sebastian Raschka # # Functions to retrieve SMILE strings from the ZINC online database # (http://zinc.docking.org) import urllib.request import urllib.parse import pyprind def get_zinc_smile(zinc_id): """ Gets the corresponding SMILE string for a ZINC ID query from the ZINC online database. Requires an internet connection. Keyword arguments: zinc_id (str): A valid ZINC ID, e.g. 'ZINC00029323' Returns the SMILE string for the corresponding ZINC ID. E.g., 'COc1cccc(c1)NC(=O)c2cccnc2' """ stripped_id = zinc_id.strip('ZINC') smile_str = None try: response = urllib.request.urlopen('http://zinc.docking.org/substance/{}'.format(stripped_id)) except urllib.error.HTTPError: print('Invalid ZINC ID {}'.format(zinc_id)) response = [] for line in response: line = line.decode(encoding='UTF-8').strip() if line.startswith('Draw')[0] smile_str = urllib.parse.unquote(line) break return smile_str def generate_zincid_smile_csv(zincid_list, out_file, print_progress_bar=False): """ Generates a CSV file of ZINC_ID,SMILE_string entries by querying the ZINC online database. Keyword arguments: zincid_list (str): Path to a UTF-8 or ASCII formatted file that contains 1 ZINC_ID per row. E.g., ZINC0000123456 ZINC0000234567 [...] out_file (str): Path to a new output CSV file that will be written. print_prgress_bar (bool): Prints a progress bar to the screen if True. """ id_smile_pairs = [] with open(zincid_list, 'r') as infile: all_lines = infile.readlines() if print_progress_bar: pbar = pyprind.ProgBar(len(all_lines), title='Downloading SMILES') for line in all_lines: line = line.strip() id_smile_pairs.append((line, get_zinc_smile(line))) if print_progress_bar: pbar.update() with open(out_file, 'w') as out: for p in id_smile_pairs: out.write('{},{}\n'.format(p[0], p[1])) def check_duplicate_smiles(zincid_list, out_file, compare_simplified_smiles=False): """ Scans a ZINC_ID,SMILE_string CSV file for duplicate SMILE strings. Keyword arguments: zincid_list (str): Path to a UTF-8 or ASCII formatted file that contains 1 ZINC_ID per row. E.g., ZINC12345678,Cc1ccc(cc1C)OCCOc2c(cc(cc2I)/C=N/n3cnnc3)OC ZINC01234567,C[C@H]1CCCC[NH+]1CC#CC(c2ccccc2)(c3ccccc3)O [...] out_file (str): Path to a new output CSV file that will be written. compare_simplified_smiles (bool): If true, SMILE strings will be simplified for the comparison. """ smile_dict = dict() with open(zincid_list, 'r') as infile: all_lines = infile.readlines() for line in all_lines: zinc_id,smile_str = line.strip().split(',') if compare_simplified_smiles: smile_str = simplify_smile(smile_str) if smile_str not in smile_dict: smile_dict[smile_str] = [zinc_id] else: smile_dict[smile_str].append(zinc_id) with open(out_file, 'w') as out: out.write('zinc_id,smile_str,duplicates') for entry in smile_dict: out.write('\n{},{},{},'.format( smile_dict[entry][0], entry, len(smile_dict[entry]) - 1)) for duplicate in smile_dict[entry][1:]: out.write(duplicate + ',') def simplify_smile(smile_str): """ Simplifies a SMILE string by removing hydrogen atoms (H), chiral specifications ('@'), charges (+ / -), '#'-characters, and square brackets ('[', ']'). Keyword Arguments: smile_str (str): A smile string, e.g., C[C@H](CCC(=O)NCCS(=O)(=O)[O-]) Returns a simplified SMILE string, e.g., CC(CCC(=O)NCCS(=O)(=O)O) """ remove_chars = ['@', '-', '+', 'H', '[', ']', '#'] stripped_smile = [] for sym in smile_str: if sym.isalpha(): sym = sym.upper() if sym not in remove_chars: stripped_smile.append(sym) return "".join(stripped_smile) ����������������������������������������������������������������������������������������������������������������������������������smilite-1.1.1/test/���������������������������������������������������������������������������������0000755�0000766�0000024�00000000000�12305163047�015235� 5����������������������������������������������������������������������������������������������������ustar �sebastian�����������������������staff���������������������������0000000�0000000������������������������������������������������������������������������������������������������������������������������������������������������������������������������smilite-1.1.1/test/test_get_zinc_smile.py�����������������������������������������������������������0000644�0000766�0000024�00000000252�12305162131�021631� 0����������������������������������������������������������������������������������������������������ustar �sebastian�����������������������staff���������������������������0000000�0000000������������������������������������������������������������������������������������������������������������������������������������������������������������������������# Sebastian Raschka, 02/2014 import smilite def test_get_zinc_smile(): out = smilite.get_zinc_smile('ZINC00029323') assert out == 'COc1cccc(c1)NC(=O)c2cccnc2' ������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������smilite-1.1.1/test/test_simplify_smile.py�����������������������������������������������������������0000644�0000766�0000024�00000000273�12305162131�021666� 0����������������������������������������������������������������������������������������������������ustar �sebastian�����������������������staff���������������������������0000000�0000000������������������������������������������������������������������������������������������������������������������������������������������������������������������������# Sebastian Raschka, 02/2014 import smilite def test_simplify_smile(): out = smilite.simplify_smile('C[C@H](CCC(=O)NCCS(=O)(=O)[O-])') assert out == 'CC(CCC(=O)NCCS(=O)(=O)O)' �����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������